Step 10. Enzyme activity assay
As cellulase is supposed to display on the surface of Pichia pastoris, we will directly conduct the enzyme activity assay using collected cells.
We take 5mL fermentation liquid from 500mL flask used for continuous fermentation and put it into a 7mL tube. Cells are then collected by centrifugation at 4℃. Supernatant in the tube are discarded, and sediment (cells) is washed by 0.05M NaAc-HAc buffer (pH=5.0) for twice. After washing, cells are resuspended by 0.05M NaAc-HAc buffer (pH=5.0) to OD600=5 for enzyme activity assay.
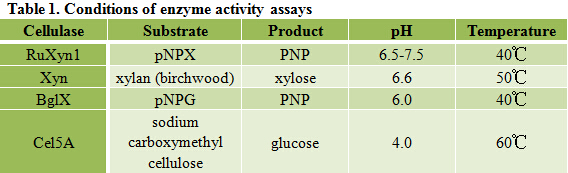
Different cellulases have different properties, including substrate specificity, optimum pH and temperature, etc, thus require different conditions for enzyme activity assays. Conditions we use for each cellulase in this project are recorded in Table 1.
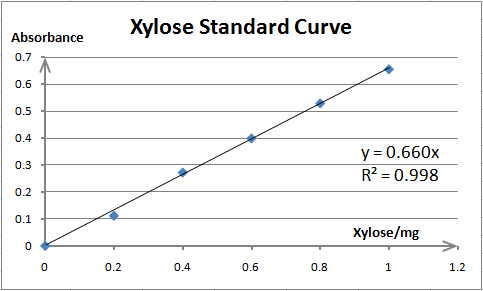
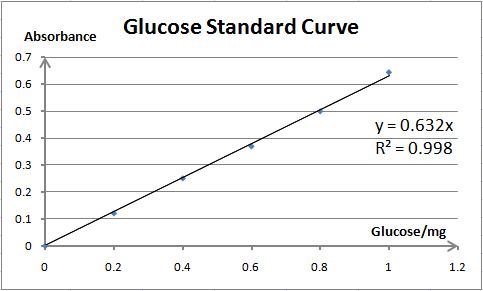
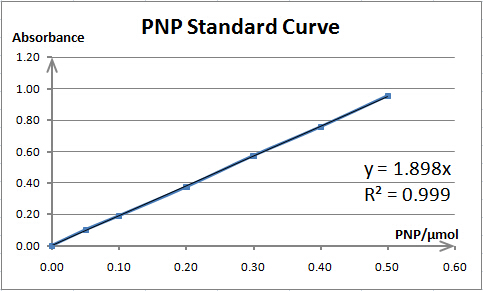
Cellulase activity is assayed by measuring the amount of product (xylose, glucose or PNP) released from corresponding substrate. The amount of product is determined by spectrophotometry. As spectrophotometry has a limited linear range, we need to make standard curves for each substrate before enzyme activity assays; thus, we can prepare samples whose concentration is qualified for reliable measurement. Also, standard curves are necessary for our converting absorbance to specific amount of product.
RuXyn1 Activity Assay
The total reaction system of 1mL contains 0.1mL enzyme solution (resuspended cells) and 0.9mL 5mmol/L pNPX (p-nitrophenyl-D-xylopyranoside) (dissolved in 0.05M NaAc-HAc buffer (pH=5.0)). For preheating, the reaction system is incubated at 40℃ for 5 minutes before the addition of enzyme solution. After incubation at 40℃ for 10 minutes, the reaction was terminated by adding 2mL 1mol/L Na2CO3. To reach a proper concentration for measurement, the reaction solution is diluted by adding ddH2O to 15mL. The RuXyn1 activity is determined by measuring the absorbance at 400nm. The background hydrolysis of the substrate is deducted by a reference sample whose content and treating process are exactly the same as the reaction mixture except no enzyme solution is added. One unit (U) of enzyme activity is defined as the amount of enzyme required for releasing 1μmol of PNP per minute.
Xyn Activity Assay
The total reaction system of 1mL contains 0.1mL enzyme solution (resuspended cells) and 0.9mL 1% xylan (birchwood) (dissolved in 0.05M NaAc-HAc buffer (pH=5.0)). The preheating, reaction and incubation processes are the same as RuXyn1 activity assay, except that the temperature is 50℃ for Xyn. After incubation, 3mL DNS is added to the reaction mixture, and the following reaction is carried out in boiling water bath. After 5 minutes of reaction, tubes are taken out from boiling water and cooled in cold water. The Xyn activity is determined by measuring the absorbance at 540nm. The background hydrolysis of the substrate is deducted by a reference sample whose content and treating process are exactly the same as the reaction mixture except no enzyme solution is added. One unit (U) of enzyme activity is defined as the amount of enzyme required for releasing 1mg of xylose per minute.
BglX Activity Assay
As BglX and RuXyn1 have the same reaction product, PNP, their activity assays are also quite similar, except that the substrate for BglX is pNPG (p-nitrophenyl-D-glucoside) (dissolved in 0.05M NaAc-HAc buffer (pH=5.0)). One unit (U) of enzyme activity is also defined as the amount of enzyme required for releasing 1μmol of PNP per minute.
Cel5A Activity Assay
As the reaction product of Cel5A and Xyn are both monosaccharide (glucose and xylose, respectively), their activity assays are also quite similar, except that the substrate for Cel5A is 1% sodium carboxymethyl cellulose (dissolved in 0.05M NaAc-HAc buffer (pH=5.0)), and the optimum temperature is 60℃. One unit (U) of enzyme activity is defined as the amount of enzyme required for releasing 1mg of glucose per minute.
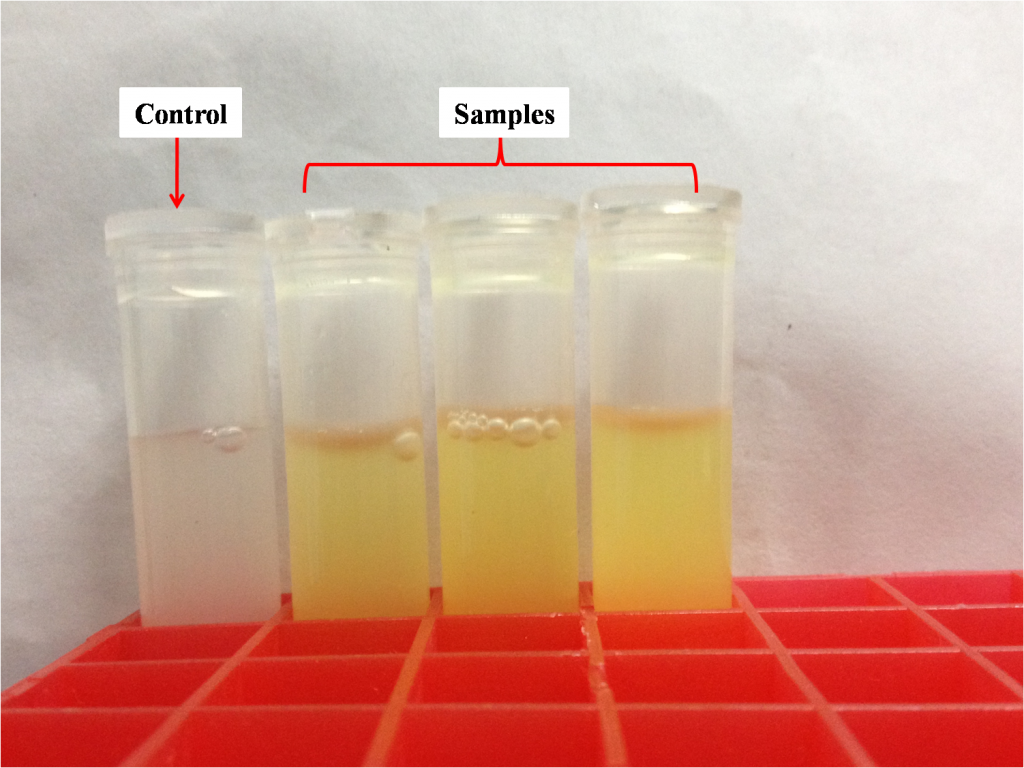
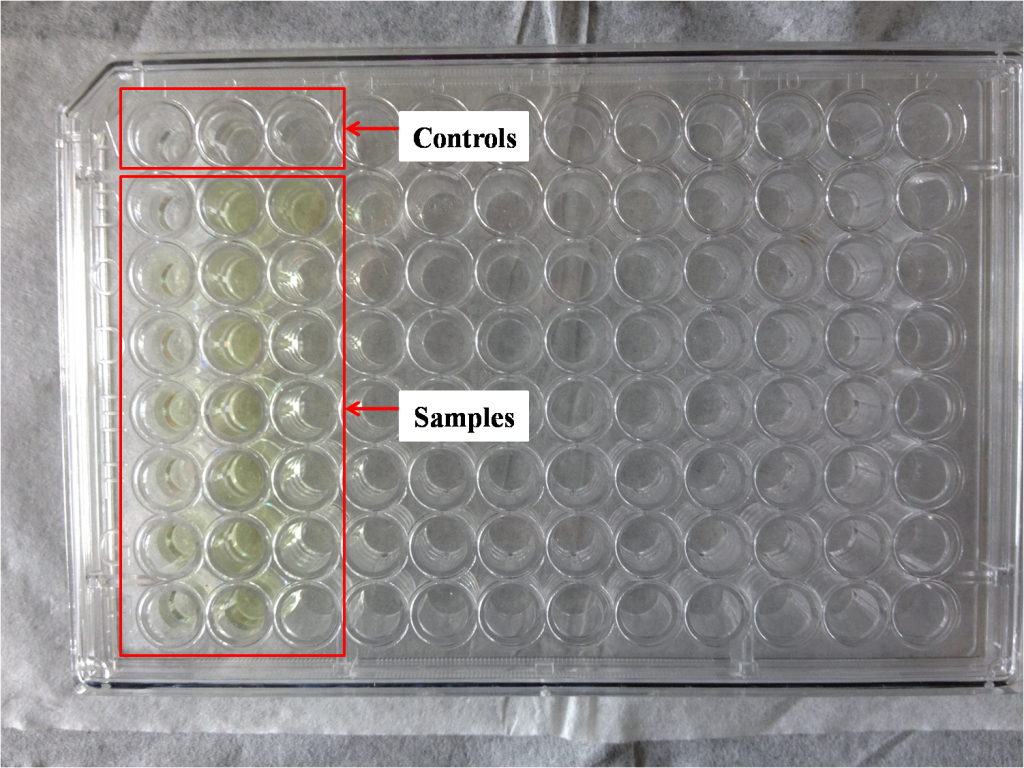
As for measuring absorbance, you can either use microplate reader or spectrophotometer. The processes of sample preparation are similar – use tubes (Figure 1) if you choose spectrophotometer, use 96-well plates (Figure 2) if you choose microplate reader.
It is easy to see that the amount of sample required for microplate reader is much fewer than that of spectrophotometer; the amount of sample required also determines the advantages and disadvantages of the two methods. Microplate reader is fast and high throughput, however has a bigger relative error. Spectrophotometer is time-consuming and the operation process is laborious, while the relative error is quite small.
After preliminary experiment, we chose spectrophotometer for our project. We have successfully obtained 9 strains carrying RuXyn1, 1 strain carrying bglX, 13 strains carrying xyn and 2 strains carrying cel5A with high enzyme activity.
How are the reagents prepared?
0.05M NaAc-HAc buffer (pH=5.0): add ddH2O to 1.43mL HAc (l) to reach a final volume of 500mL, and add NaOH (s) to pH=5.0
DNS: sequentially add 10.00g DNS, 16.00g NaOH, 5.00g Phenol, 5.00g Na2SO3 and 300.00g C4O6H4KNa to 750mL ddH2O preheated at 45℃, and add ddH2O to 1000mL when completely dissolved. Store the reagent in a brown reagent bottle and place it at room temperature for at least 7 days before use.