Step 3. Determine the sequence of the plasmid by sequencing
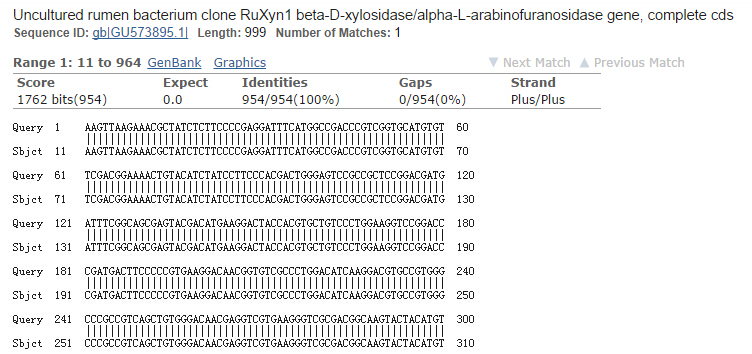
There’s a chance of gene mutation during the process of vector construction. The precise order of nucleotides within our vector is determined by DNA sequencing. Plasmid samples are sent to TSINGKE Biological Technology (other companies with sequencing services are also okay). We get reports of the sequences of our samples; and use BLAST to do sequence alignment.
Our BLAST result of RuXyn1 is shown in the image above, indicating that the sequence of RuXyn1 in our sample is correct. The sequencing results of the other three enzyme genes (Xyn, BglX, Cel5A) are processed in the same way. If the sequencing result shows that the gene mutated, the vector should be reconstructed until you get the correct sequence.
Step 4. Linearize the plasmid to stimulate recombination
Linearization is to use restriction enzyme to digest and linearize the circular plasmid. Linearized DNA can generate stable transformants of Pichia pastoris via homologous recombination between the transforming DNA and regions of homology within the genome. Such integrants show extreme stability in the absence of selective pressure even when present as multiple copies.
The type of restriction enzyme used in linearization is determined both by the type of the host cell (Pichia Pastoris strain GS115 or KM71) and the phenotype of transformants (His+ Mut+ or His+ Muts). His+ represents the “Histidine synthesizing” phenotype, Mut+ represents the “Methanol utilizing” phenotype and Muts refers to the “Methanol utilization slow” phenotype.
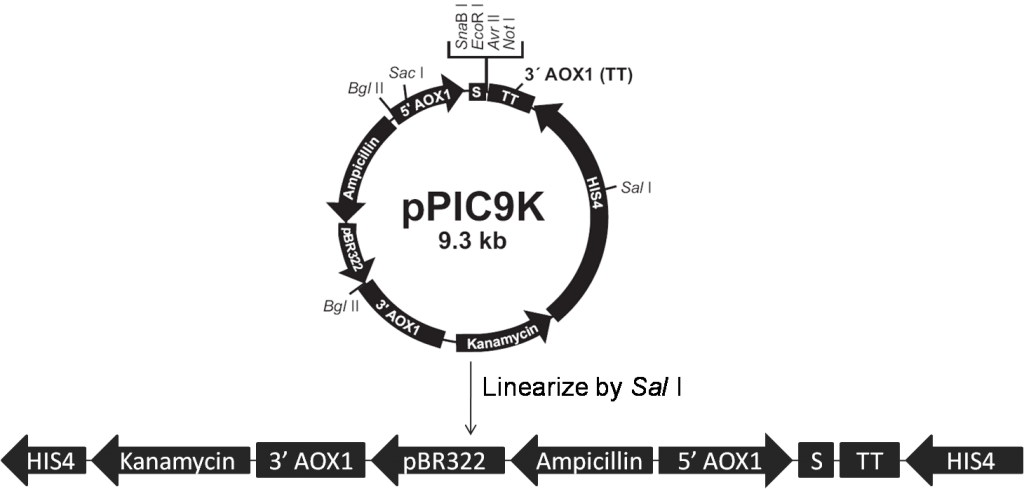
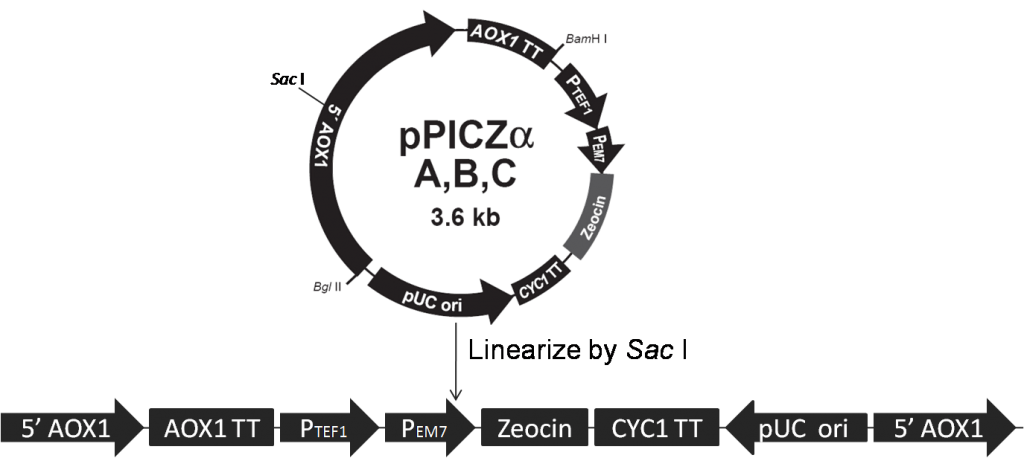
The Pichia Pastoris strain GS115 is used as host cell in our project. GS115 has a mutation in its histidinol dehydrogenase gene (his4) that prevents it from synthesizing histidine (His–). Both pPIC9K and pPICZα contain the HIS4 gene that complements his4 in the host cell (His+), thus transformants are selected for their ability to grow on histidine-deficient medium. We use Sal I and Sac I to linearize our expression vectors, pPIC9K (Figure 1) and pPICZα (Figure 2) respectively, according to Pichia Expression Kit (page32, Invitrogen).
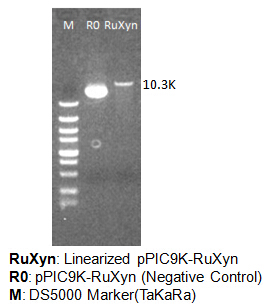
Linearized plasmids are identified by agarose gel electrophoresis. Circular DNA runs faster than linearized DNA with the same primary structure. Here I take the linearization of pPIC9K-RuXyn as an example (Figure 3). We use the plasmid without enzyme digestion (pPIC9K-RuXyn) as negative control (R0), which runs ahead of the linearized DNA (RuXyn) in the agarose gel.
Step 5. Electroporation for the first round
Electroporation is to use electrical field to introduce the expression vector into the yeast cell. One kind of expression vector is introduced into the cell in one round of electroporation. Two rounds of electroporation are required, for we want to co-transform two enzyme genes into Pichia pastoris and co-display the two enzymes.
We use the method in Pichia Expression Kit (page 77, Invitrogen) to prepare electro competent cells and electroporation. pPIC9K-RuXyn, pPIC9K-BglX, pPICZα-Xyn and pPICZα-Cel5A are introduced into P. pastoris GS115 to construct GS115-9K-RuXyn, GS115-9K-BglX, GS115-Zα-Xyn and GS115-Zα-Cel5A. pPIC9K and pPICZα, both carry Sed1p only, are also introduced into P. pastoris GS115 to construct GS115-9K and GS115-Zα, which function as the negative control for Immunofluorescence Assay and Flow Cytometry analysis.