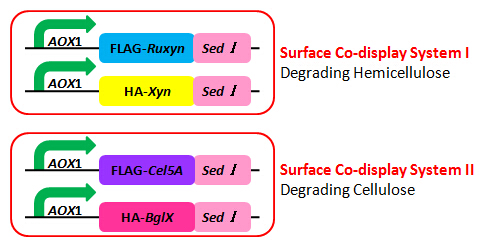
Among all the cellulases targeting different chemical bonds in lignocellulose, our group found two sets with synergy in the literature. Synergy in this context is a property that the two enzymes can enhance the activity of each other when functioning together. The first set is RuXyn1 (beta-D-xylosidase) and Xyn (beta-xylanase); and the second set is BglX (beta-glucosidase) and Cel5A (endoglucanase). The two sets of cellulases are responsible for degrading hemicellulose and cellulose respectively.
As for lignin, an aromatic polymer, pretreatment is required to remove this lignin from cellulose and hemicellulose. Without the protection of lignin, cellulose and hemicellulose will lose their resistance to enzymes and can be more easily degraded. Previous studies gave many options of enzymes used for pretreatment, such as Lip (Lignin peroxidase), MnP (Mn peroxidase) and GLOX (Glyoxal oxidase). So, our project focused on the degradation of cellulose and hemicellulose.
The process of constructing the surface co-display system is as follows.
Step 1. Choose a proper host cell and the corresponding anchor protein
As cellulases are all originated from fungi, we need a eukaryotic expression system. Pichia pastoris and Saccharomyces cerevisiae are both modern frequently used eukaryotic expression systems. We chose Pichia pastoris in our project, because it has several advantages over Saccharomyces cerevisiae: 1) it’s modification ability is stronger, capable of peptide folding, glycosylation, methylation and acetylation; 2) it contains promoter PAOX1, the regulation of which is the most efficient and strictest among the promoters discovered currently; 3) exogenous genes transformed into Pichia pastoris integrate into its genome, expressing steadily; while genes transformed into Saccharomyces cerevisiae exist as plasmid that is very likely to be rejected by the cell after several generations.
pPIC9K and pPICZα are both good expression vectors in Pichia pastoris, but not all the cells transformed with the vector can successfully express the target protein on its surface. To reach a high activity of our whole-cell catalyst, we want the display ratio (the amount of cells expressing the target protein on the surface/ total cells) as high as possible, which depends on both the expression vector and the anchor protein. In our lab, the accessible candidates are Cwp2p, Sed1p and α-agglutinin. We expressed the candidates fused with EGFP (Enhanced Green Fluorescent Protein) based on pPIC9K and pPICZα respectively; and use FCM (Flow Cytometry) to test their display ratio. From our experiments we found that Sed1p is the best, with the display ratio of 99.50% in pPIC9k and 98.36% in pPICZα.
Step 2. Construct the expression vectors
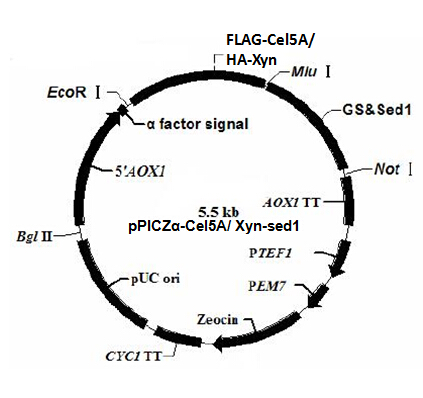
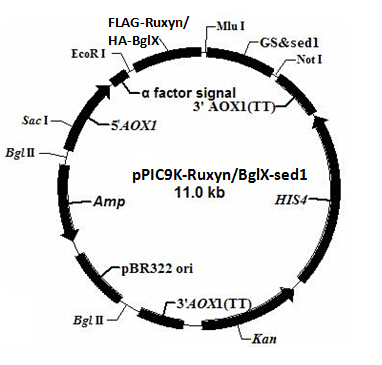
The plasmid profiles are shown in figure 1 and 2. α factor signal is a secretion signal; PAOX1 is an induced promoter (functions only in the presence of specific inductor) sensitive to methanol. EcoR I, Mlu I and Not I are restriction sites used for the vector construction. We add a tag (FLAG tag or HA tag) to the target gene for the detection of the target protein.
Tags have their specific primary antibody. The primary antibody can be recognized by the secondary antibody through antigen-antibody reaction. The secondary antigen is labeled by fluorescent. Thus we can detect the target protein by detecting the fluorescence on the cell surface under the fluorescence microscope. To distinguish the two target proteins, we use two different tags (FLAG tag and HA tag). Different tags correspond to different primary and secondary antibodies, emitting fluorescence of different color.
Figure 3 is the schematic of the two surface co-display systems. pPIC9K has resistance against ampicillin and kanamycin, while pPICZα is resistant to zeocin. Notice that two enzyme genes co-expressed in one cell type are constructed in two different plasmid backbones; we can easily obtain the strain with both two expression vectors by resistance screening assay.
According to the plasmid backbone and the inserted gene, we name the four expression vectors pPIC9K-RuXyn, pPIC9K-BglX, pPICZα-Xyn and pPICZα-Cel5A.